In the second year of the project, we deposited in the international GenBank database information on 20 new nucleotide sequences of fungi whose fruiting bodies were collected directly in the sites where sampling for metagenomic analysis was performed. The complete nucleotide sequences of ITS1-5.8S-ITS2 nrDNA obtained and deposited in NCBI GenBank are represented mainly by taxa about which there is no information in reference databases. In particular, information on the primary structure of the ITS region of nrDNA was obtained for the first time for corticioid fungi from the genera Peniophorella (Hymenochaetales), Hyphoderma, Phlebia (Polyporales), and Amylocorticium (Amylocorticiales). In addition, nucleotide sequences were obtained from authentic samples for little-known and rarely found in nature representatives of the genera of xylotrophic basidiomycetes Athelia (Atheliales), Leucogyrophana (Boletales), Kneiffiella (Hymenochaetales), Yuchengia (Polyporales). Information on 146 fungal findings from a mass windthrow area in the mixed broadleaved forest was published in GBIF.
A total of 4717 different ITS1 sequence variants of fungi and 92262 sequence variants of the V4 regions of the 16S rRNA gene of bacteria were obtained from deadwood metagenomes. After downsampling, about 2500 and about 22500 unique sequences remained in the fungal and bacterial community, respectively (with only 643 and 665 of these representing one per cent or more of all community ASVs in at least one sample).
All tree species examined showed an increase in the alpha diversity of the fungal and bacterial communities between the initial and final stages of decomposition, with the bacterial community being more diverse than the fungal community at all stages of decomposition. In the first stages, the fungal community consisted on average of about 25 species with strongly dominant representatives. The exception was Picea abies, on which more than 60 species were found. The bacterial community of each tree species consisted of over 200 species, except for Acer platanoides and Betula pendula, in which 45 and 86 species were observed, respectively. By the fourth and fifth stages of decomposition, an average of over 100 fungal species and between approximately 400 and over 600 bacterial species were observed in all tree species. Some tree species showed a decline in the diversity of the investigated destructors at the second or third stage of decomposition. With the overall high noisiness of the data, the concentration of amplicons from the bacterial community was more often higher at the last stage of decomposition than at the first, whereas the fungal community often showed a peak at earlier stages (1-3) and a decline at the last. According to our data, the highest concentration of amplicons from fungi and bacteria was characteristic for Tilia cordata and the lowest for Quercus rubra. In oak wood, bacteria appeared in noticeable numbers only by the third stage of decomposition, which probably correlates with its resistance to decomposition and, consequently, its active use in the economy. Interestingly, in some cases (e.g. at the first stage of decomposition in Fraxinus excelsior and the second in Picea) fungi significantly prevailed over bacteria, which may be due to the release of bactericidal substances by them, and the number of bacteria increased with the transition to later stages of decomposition.
The fungal community was characterised by very high beta diversity (mean Bray-Curtis distance 0.96 ± 0.08), with the distance between samples of different tree species of the same decomposition stage being slightly smaller than the distance between samples of the same species but different stages, resulting in a more pronounced clustering of samples by decomposition stage than by tree species. During wood decomposition, the fungal community for some tree species changed dramatically, showing a change of dominant representatives at the phylum level. Of the 47 classes found in the fungal community, 13 were observed in at least one sample of each species and each stage of decomposition. At the same time, most fungal genera were observed only on samples of one species of one stage, suggesting that there is no single common pathway of wood decomposition by the fungal community. In the bacterial community, 13 out of 104 classes and 146 out of 781 genera were observed in samples of all types, and, as in the fungal community, frequent classes included dominant representatives. When beta diversity was assessed using the Bray-Curtis distance at the ASV level, bacterial communities also varied greatly between samples. In terms of similarity of bacterial communities (as with fungi, but more so for bacteria), samples clustered more by stage than by tree species (Figure).
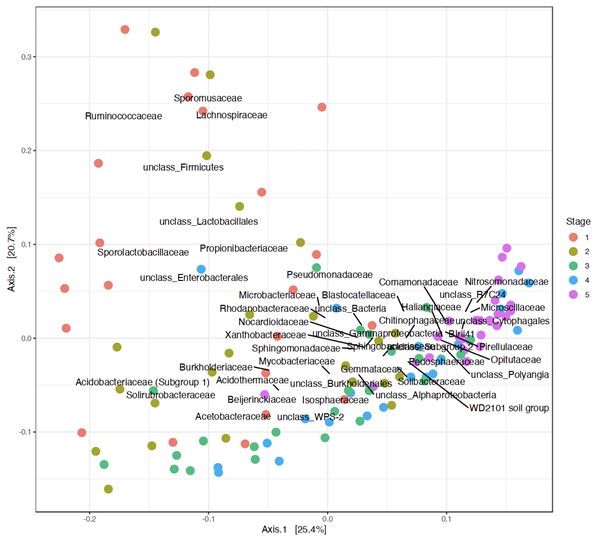
Figure. Ordination of bacterial communities of samples using the Principal coordinate method (PCoA). The figure shows samples of all studied tree species. Different colors indicate the stages of decomposition. Percentages of explained variance are marked on the axes. (From Volobuev et al. Proceedings of 11th Moscow Conference on Computational Molecular Biology MCCMB'23. Moscow.2023) https://mccmb.belozersky.msu.ru/2021/thesis/MCCMB.utf8.html
Based on the records of fruiting bodies of aphyllophoroid fungi, 127 species of basidial fungi were identified on dead tree trunks of 8 tree species. The fungal species belonged to 78 genera, 50 families, and 14 orders. The maximum number of fungal species (46) occurred on Picea and minimum (11) on Quercus while 38, 24, 22, 19, 18, and 13 were surveyed on Acer, Fraxinus, Populus tremula, Ulmus glabra, and Tilia, respectively. White rot fungi dominated (74%), while brown rot fungi accounted for 19%; morphologically, corticoid and popypore comprised 55 and 32%, respectively, and clavarioid and hydnoid together accounted for 12% of sporocarps. The mean number of fungal species was 1.8 ± 0.06 in the 1-m plot and 10.4 ± 0.98 in the trunk. High gamma diversity was supported by high beta diversity of the fungal community: mean Bray-Curtis distance was 0.95 ± 0.001 and 0.92 ± 0.006 for plots and trunks, respectively. Trunks of different tree species did not differ in the number of fungal species found on them, while groups of tree species differed significantly: the number of fungal species found on ring-porous angiosperms (10.6 ± 1.2) was significantly lower than those found on the gymnosperm Picea (16.8 ± 2.6); Kruskal–Wallis test χ2 = 9.48, P = 0.009. The composition of basidiomycetes on lying trunks of various tree species differed rather well, as shown by the results of non-metric multidimensional scaling (NMDS) and permutational multivariate analysis (PERMANOVA). Tree species identity determined 45% of the variation in basidiomycete species composition (P = 0.0001); together with mean stem decomposition stage (R2 = 5%, P = 0.05), dbh and number of non-zero sites, the model determined 58% of the variation in fungal species composition. Additive partitioning of biodiversity showed that the contributions of all diversity components were significant, but the maximum contribution (81%) to the total gamma diversity of basidiomycetes was made by compositional diversity among tree species, which was significantly higher than that expected under the null model.
Thus, we obtained overall consistent estimates of the diversity of fungal communities identified from fruiting bodies and metagenomic analyses. The two analyses showed an increase in community diversity with increasing decomposition stage and the importance of the contribution of woody substrate diversity to the total gamma diversity of the fungal population of the analyzed site. Metagenomic analysis allowed a detailed assessment of the quantitative and qualitative change in the species diversity of destructor communities (bacteria and fungi) during wood decomposition in different tree species; specific features of the decomposition process for each of the eight tree species analyzed were suggested.
The results of the research were presented at 4 scientific conferences. Nine scientific articles were published, among them one in Q1 journal (IF 4.384); one in a journal registered in Scopus database; also 4 articles in conference proceedings and 2 datasets were published.



